Building a web app - Part 1 Supporting tagline
I’d like to start off this series of posts by taking a look at the data we’re going to be working with. As a reminder, I’m sharing the code for this series at https://github.com/dkincaid/web-app-explore.
As I mentioned last post the data that we’re going to use comes from the Tycho project at University of Pittsburgh. I’m going to use the Level 2 data. I chose this data for a couple reasons. The first is that it is a little larger dataset (47 diseases over 1888-2013) and offers an API to access the data.
For more information on this data see:
Willem G. van Panhuis, John Grefenstette, Su Yon Jung, Nian Shong Chok, Anne Cross, Heather Eng, Bruce Y Lee, Vladimir Zadorozhny, Shawn Brown, Derek Cummings, Donald S. Burke. Contagious Diseases in the United States from 1888 to the present. NEJM 2013; 369(22): 2152-2158.
Overview
I have already downloaded and processed all the data for all the states into a file named
states_cases.Rda. Let’s load that up and have a look.
options(width = 120, warn = -1)
load("states_cases.Rda")
str(states_cases)
## 'data.frame': 819885 obs. of 6 variables:
## $ year : chr "1942" "1942" "1942" "1942" ...
## $ week : chr "1" "2" "3" "4" ...
## $ state : Factor w/ 57 levels "AL","AR","AZ",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ number : int 0 0 0 0 0 0 0 0 0 0 ...
## $ event : Factor w/ 1 level "CASES": 1 1 1 1 1 1 1 1 1 1 ...
## $ disease: Factor w/ 39 levels "ANTHRAX","BOTULISM",..: 1 1 1 1 1 1 1 1 1 1 ...
summary(states_cases)
## year week state number event
## Length:819885 Length:819885 NY : 82686 Min. : 0 CASES:819885
## Class :character Class :character CA : 22953 1st Qu.: 1
## Mode :character Mode :character TX : 21116 Median : 5
## OH : 20316 Mean : 65
## FL : 20070 3rd Qu.: 28
## MI : 19188 Max. :89363
## (Other):633556
## disease
## MEASLES :118727
## WHOOPING COUGH [PERTUSSIS] : 75424
## DIPHTHERIA : 65007
## SCARLET FEVER : 64310
## SMALLPOX : 53461
## TYPHOID FEVER [ENTERIC FEVER]: 41711
## (Other) :401245
So we have counts of the number of cases reported in each state for each week of the year. What is the range of years?
c(min(states_cases$year), max(states_cases$year))
## [1] "1900" "2013"
Let’s see how the total cases for each disease in the data. Here I’m going to use the new package
from Hadley Wickam, dplyr (which I am loving!).
library(dplyr, quietly = TRUE, warn.conflicts = FALSE)
states_cases %.% group_by(disease) %.% summarise(total = sum(number)) %.% arrange(desc(total))
## Source: local data frame [39 x 2]
##
## disease total
## 1 MEASLES 22591587
## 2 INFLUENZA 6741366
## 3 CHLAMYDIA 5237336
## 4 SCARLET FEVER 5166016
## 5 GONORRHEA 4117671
## 6 WHOOPING COUGH [PERTUSSIS] 2813601
## 7 CHICKENPOX [VARICELLA] 1808532
## 8 MUMPS 914045
## 9 DIPHTHERIA 910819
## 10 STREPTOCOCCAL SORE THROAT 638494
## 11 RUBELLA 479160
## 12 TUBERCULOSIS [PHTHISIS PULMONALIS] 300760
## 13 PNEUMONIA 286079
## 14 TYPHOID FEVER [ENTERIC FEVER] 259413
## 15 SMALLPOX 242003
## 16 SALMONELLOSIS 205408
## 17 RABIES IN ANIMALS 148768
## 18 LYME DISEASE 128033
## 19 GIARDIASIS 96853
## 20 SHIGELLOSIS 68185
## 21 DYSENTERY 56341
## 22 MALARIA 37629
## 23 BRUCELLOSIS [UNDULANT FEVER] 35940
## 24 COCCIDIOIDOMYCOSIS 34481
## 25 CRYPTOSPORIDIOSIS 30581
## 26 ROCKY MOUNTAIN SPOTTED FEVER 28198
## 27 LEGIONELLOSIS 20464
## 28 TULAREMIA 17088
## 29 TYPHUS FEVER 16220
## 30 MENINGITIS 4456
## 31 TETANUS 2639
## 32 TRICHINIASIS 2027
## 33 TOXIC SHOCK SYNDROME 1957
## 34 PSITTACOSIS 1345
## 35 ANTHRAX 204
## 36 LEPROSY 150
## 37 POLIOMYELITIS 64
## 38 BOTULISM 15
## 39 PELLAGRA 0
and what about the top 10 years?
states_cases %.% group_by(year) %.% summarise(total = sum(number)) %.% arrange(desc(total)) %.% head(10)
## Source: local data frame [10 x 2]
##
## year total
## 1 1941 2063853
## 2 1928 1755548
## 3 1938 1738608
## 4 1943 1527009
## 5 1939 1490709
## 6 1929 1381464
## 7 1935 1359174
## 8 1946 1358645
## 9 1944 1344990
## 10 1973 1264263
How about looking at the worst 10 years for each disease (I’ll just show the first 3 diseases here to save some space)
topyear <- function(d) {
subset(states_cases, disease == d) %.% group_by(year) %.% summarise(total = sum(number)) %.% arrange(desc(total)) %.% head(10)
}
topyears.by.disease <- lapply(levels(states_cases$disease), topyear)
names(topyears.by.disease) <- levels(states_cases$disease)
head(topyears.by.disease, 3)
## $ANTHRAX
## Source: local data frame [4 x 2]
##
## year total
## 1 1943 90
## 2 1942 67
## 3 1944 45
## 4 1945 2
##
## $BOTULISM
## Source: local data frame [1 x 2]
##
## year total
## 1 1952 15
##
## $`BRUCELLOSIS [UNDULANT FEVER]`
## Source: local data frame [10 x 2]
##
## year total
## 1 1947 6436
## 2 1946 5830
## 3 1945 4623
## 4 1948 2631
## 5 1952 2098
## 6 1953 1823
## 7 1954 1812
## 8 1955 1266
## 9 1956 1091
## 10 1957 915
So we can see that for each disease we have very different amounts of data. Let’s see what the min and max year is for each disease
states_cases %.% group_by(disease) %.% summarise(min = min(year), max = max(year), range = as.numeric(max) - as.numeric(min)) %.% arrange(disease)
## Source: local data frame [39 x 4]
##
## disease min max range
## 1 ANTHRAX 1942 1945 3
## 2 BOTULISM 1952 1952 0
## 3 BRUCELLOSIS [UNDULANT FEVER] 1945 1981 36
## 4 CHICKENPOX [VARICELLA] 1972 2013 41
## 5 CHLAMYDIA 2006 2013 7
## 6 COCCIDIOIDOMYCOSIS 2006 2013 7
## 7 CRYPTOSPORIDIOSIS 2006 2013 7
## 8 DIPHTHERIA 1920 1981 61
## 9 DYSENTERY 1942 1948 6
## 10 GIARDIASIS 2006 2013 7
## 11 GONORRHEA 1972 2013 41
## 12 INFLUENZA 1919 1951 32
## 13 LEGIONELLOSIS 1982 2013 31
## 14 LEPROSY 1942 1945 3
## 15 LYME DISEASE 2006 2013 7
## 16 MALARIA 1952 2013 61
## 17 MEASLES 1909 1982 73
## 18 MENINGITIS 1917 1959 42
## 19 MUMPS 1968 2013 45
## 20 PELLAGRA 1928 1929 1
## 21 PNEUMONIA 1920 1951 31
## 22 POLIOMYELITIS 1939 1939 0
## 23 PSITTACOSIS 1956 1961 5
## 24 RABIES IN ANIMALS 1948 2013 65
## 25 ROCKY MOUNTAIN SPOTTED FEVER 1942 2011 69
## 26 RUBELLA 1966 2001 35
## 27 SALMONELLOSIS 2006 2013 7
## 28 SCARLET FEVER 1909 1966 57
## 29 SHIGELLOSIS 2006 2013 7
## 30 SMALLPOX 1900 1952 52
## 31 STREPTOCOCCAL SORE THROAT 1960 1961 1
## 32 TETANUS 1962 1972 10
## 33 TOXIC SHOCK SYNDROME 1983 1994 11
## 34 TRICHINIASIS 1952 1955 3
## 35 TUBERCULOSIS [PHTHISIS PULMONALIS] 1921 2013 92
## 36 TULAREMIA 1942 1973 31
## 37 TYPHOID FEVER [ENTERIC FEVER] 1909 1983 74
## 38 TYPHUS FEVER 1942 1947 5
## 39 WHOOPING COUGH [PERTUSSIS] 1909 2013 104
Graphical visualizations
Finally, let’s look at a couple of plots. Here is a look at diphtheria and whooping cough in the states of New York, California, Texas and Illinois.
library(ggplot2, quietly = TRUE)
di <- c("DIPHTHERIA", "WHOOPING COUGH [PERTUSSIS]")
dip_whoop <- subset(states_cases, disease %in% di & state %in% c("NY", "CA", "TX", "IL"))
ggplot(dip_whoop, aes(x = as.Date(paste("1", week, year, sep = "-"), format = "%w-%W-%Y"), y = number)) + geom_bar(stat = "identity", width = 7) + scale_y_continuous("Cases", limits = c(0,
1500)) + scale_x_date("Week") + facet_grid(disease ~ state)
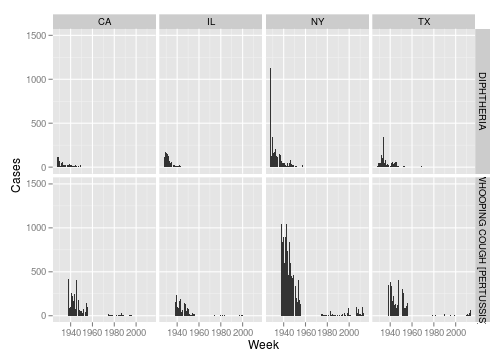
Now I’ll create a matrix for Whooping Cough with states as rows and years as columns and create a heatmap.
library(reshape2, quietly = TRUE, warn.conflicts = FALSE)
library(RColorBrewer, quietly = TRUE, warn.conflicts = FALSE)
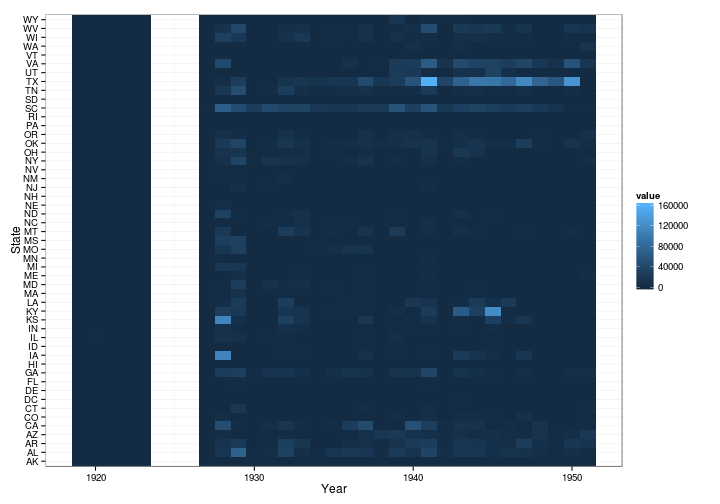
influenza.year.by.state <- acast(subset(states_cases, disease == "INFLUENZA"), state ~ year, value.var = "number", fun.aggregate = sum)
influenza.year.by.state[1:10, 1:10]
## 1919 1920 1921 1922 1923 1927 1928 1929 1930 1931
## AL 15 0 0 0 0 416 15705 69455 3281 5925
## AR 213 88 0 0 0 404 12323 20058 2529 3504
## AZ 0 0 0 0 0 1 4347 628 348 462
## CA 182 288 0 0 0 112 44906 5398 1716 5987
## CO 0 0 0 0 0 2 5677 998 11 0
## CT 60 15 0 0 0 49 1068 16748 251 1300
## DC 0 126 0 0 0 0 571 2028 36 219
## DE 14 1 0 0 0 3 43 486 18 275
## FL 114 12 0 0 0 39 1614 3737 104 1496
## GA 204 54 0 0 0 467 23688 29033 3348 12325
m <- melt(influenza.year.by.state)
ggplot(m, aes(x = Var2, y = Var1, fill = value)) + geom_tile() + scale_colour_brewer() + labs(x = "Year", y = "State", color = "Cases") + theme_bw()
Next time
I think that is enough for now. That should give you a decent sense for the data we’ll be working with. Next time I’ll start creating the Shiny application that will let someone do some of this exploration on a web based UI.
blog comments powered by Disqus